Copyright 2012 neutronsources.org | All rights reserved. | Powered by FRM II | Imprint / Privacy Policy
Following protein aggregation in real time by neutron spectroscopy
Date: 19 August 2020
Source: ILL
Protein aggregation into amyloid superstructures is the molecular manifestation of a large variety of neurodegenerative diseases such as Alzheimer’s and Parkinson’s. Evidence is increasing that transient on-pathway oligomers are actually the toxic species, such that time-resolved monitoring of protein aggregation is highly desirable.
Whereas time-resolved structural techniques have been developed and applied to study protein aggregation, methods accessing protein center-of-mass and internal diffusion dynamics remain only sparsely available.
Yet, changes in protein dynamics have been postulated to play an essential role in driving and accompanying protein aggregation. Scientists of the Institut Laue Langevin, the Institut de Biologie Structurale and the University of Copenhagen developed a time-resolved version of incoherent neutron scattering experiments on IN16B to follow protein aggregation and applied it to study the assembly of lysozyme in aqueous solution into particulate superstructures.
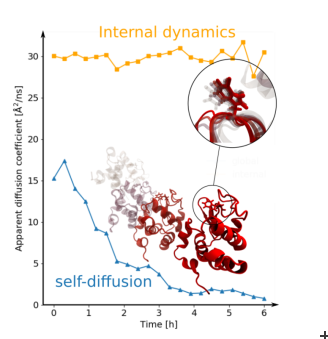
Surprisingly, the internal protein dynamics on the nano-to picosecond time scale does not change during the entire aggregation process. The center-of-mass diffusion, on the other hand, decreases as aggregation proceeds and can be well explained by a single exponential process.
By complementing the neutron results with fluorescence measurements, electron microscopy, infrared spectroscopy, x-ray powder diffraction, and dynamic light scattering, a comprehensive picture was painted in which lysozyme particulate formation is a one-step process with protein backbone and side chain dynamics remaining unchanged throughout aggregation.
The work establishes a framework to follow protein aggregation quantitatively in real time at a molecular level, simultaneously accessing center-of-mass and internal diffusivities, which will be invaluable for addressing pathological pathways of protein aggregation. This framework is not limited to proteins, but can be applied to macromolecules in general to study a large variety of processes such as self-assembly and emerging aggregates and crystals.