Copyright 2012 neutronsources.org | All rights reserved. | Powered by FRM II | Imprint / Privacy Policy
Neutrons reveal hidden secrets of the hepatitis C virus
Date: 16/01/2018
Source: www.ill.eu
p7 is a protein essential for the release of the hepatitis C virus, however little data is currently available on the way it interacts with its environment, hindering the development of vaccinations for it. Researchers have observed the structure of a functional p7 protein within its native environment for the first time using reflected neutrons. The specific protein insertion mechanism observed will help to outline potential target mechanisms for future drug development.
The hepatitis C virus (HCV) is a blood born virus that causes liver disease and cancer, with more than 300,000 people dying each year and 71 million people living with a chronic infection worldwide . While antiviral medicines are currently used, there is no vaccination currently available and side effects can results in a wrong diagnosis.
In the search to find novel therapies for HCV, researchers have looked to the membrane protein p7, which plays a key role in the release of the virus, for answers. However, there is little data available, and the crystallographic structure of the protein has not yet been resolved.
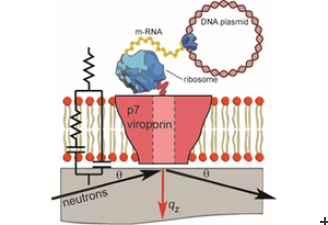
Recent investigations using neutrons have led to the development of a novel method to study the protein’s integration and structure within a native biological membrane environment. A collaboration between Synthelis SAS, University Grenoble Alpes, and the Institute Laue-Langevin (ILL) enabled researchers to observe the structure of a functional p7 protein complex from HCV for the first time within a physiologically relevant lipid bilayer, at nanoscale resolution.
To do this, the scientists performed neutron reflectometry (NR) on FIGARO, a time of flight reflectometer at the world’s flagship centre for neutron science, ILL in Grenoble, France. Momentum transfer ranges of 0.008> qz> 0.2 Å-1 and minimum reflectivities of R ~ 5×10-7 were measured using wavelengths λ = 2-20 Å, two angles of incidence and a dqz/qz resolution of 10%.
The Nature Scientific Reports study found that the p7 protein from HCV assembles within the lipid membrane into oligomers that take the shape of a funnel. The conical shape indicates a preferred protein orientation, revealing a specific protein insertion mechanism, and helping to outline potential target mechanisms for future drug development.
As membrane protein dysfunction is also correlated with a wide range of diseases, this advancement in methods to analyse membrane proteins in their native condition, at an atomic scale, also has the potential to help support new therapeutic approaches in other areas, such as for the development of antibodies against HIV.
Erik Watkins, former ILL FIGARO Instrument Scientist, said: “This new approach is a simple and efficient method complementary to other structural and more complex techniques such as NMR and crystallography. This has proved a powerful tool for characterising the protein conformation in its natural environment and one we can look to use for membrane protein discoveries not just in advancements in HCV, but further afield as well.”
Bruno Tillier, Managing Director, Synthelis added: “Neutrons have proved a key resource for this project as we needed to analyse the p7 protein structure in a specific environment. Now we can look to take this deep understanding of the virus not only to devices, like biosensors, but also to study the behaviour of membrane proteins in lipid bilayers to other fields.”
Donald Martin, Head of the research team SyNaBi and professor at University Grenoble Alpes also said: “These new results augur well for our continued development of novel nanostructured systems and devices. The ongoing fruitful collaboration between physicists, biologists and engineers from these institutions in Grenoble provides the important fundamental understanding of physical and biological processes that underlies the development of such nanostructured systems and devices.”
Thomas Soranzo, University Grenoble Alpes and former Synthelis scientist also said: “a major bottleneck in neutron reflectivity analysis of membrane proteins in planar bilayer is the sufficient insertion of polypeptides. This combinatory, new method not only allows significant incorporation of material but also allows specific labelling that could improve membrane protein structure/function studies.”
Original Publication
T. Soranzo, D. K. Martin, J.-L. Lenormand, E. B. Watkins
Coupling neutron reflectivity with cell-free protein synthesis to probe membrane protein structure in supported bilayers
Scientific Reports (2017)
DOI: 10.1038/s41598-017-03472-8